It is crucial for publication to ensure that the genes selected for regression analysis were chosen on objective criteria. This requires the reproduction of the set of genes that were called monoallelic in the manuscript drafted by Ifat. I’ll refer to that in this document as the previous manuscript. If perfect reproduction fails then minimum discordance is desired, and the regression analysis will potentially need to be repeated accordingly with the new set of genes called monoallelic.
This post was extended with the test for reference/nonreference bias, whose results are summarized in ref-nonref-test.csv for top scoring genes.
Procedures from the previous manuscript
Filters
The previous manuscript makes several statements on how the data were filtered before the gene were ranked and presented on Fig 1:
section Methods: Allelic expression analyses, p3
there had to be ≥7 reads, with a quality score ≥20; a further gene-wide filter requires 20 or more such reads for the gene to be assessable in a given individual
section Methods: Allelic expression analyses, p4
Genes that were supported by fewer than 25 subjects were excluded.
section Results: Pipeline, p4
there had to be ≥7 reads; a further gene-wide filter requires 15 or more such reads for the gene to be assessable in a given individual
section Results: Landscape of monoallelic expression[…], p6
sufficient coverage defined as >20 reads mapping to heterozygous SNPs per individual in at least ten individuals
In addition, for the regression analysis a stronger filtering was used:
section Results: Relaxation of imprinting, p8
We examined genes where we had greater than 180 analyzable individuals and where 30% or more of those individuals displayed monoallelic expression defined by Sg >0.9. An analyzable individual was defined as one with at least 50 reads at one or more SNPs imputed to be heterozygous (Lhet cutoff of 0.95 as above).
My interpretation
Two kind of filters were used: I call them read count-based and individual-based. These have the following properties:
- the read count-based filter removes any such pair (i,g) of individual i and genes g for which the total read count nig<trc, where trc is what I call a read count threshold
- Note that there is another read count-based filter, which removes pairs (s,g) of SNP s and gene g in the same fashion as the previous one but with threshold 7. However, I cannot investigate this filter until the SNP-wise read counts become easily accessible.
- the individual-based filter removes any genes g (across all individuals) if read count data involving g are available on less than tind number of individuals
Note that in fact there are two read count-based filters: not only the one for individual SNPs and a
Given the above quotations from the previous manuscript the read count-based filter might have been applied with trc=20 or 15 or 21, whereas the individual-based filter with tind=25 or 10 in order to arrive at Fig 1.
Ranking and calling monoallelic expression
The ranking of genes is based on the following score. The score of gene g is the fractions of individuals i for whom sig>0.9. In other words the score of g is based on the empirical cumulative distribution function ECDF ˆFg or, equivalently, on the survival function 1−ˆFg: the score of gene g=1−ˆFg(0.9).
Judged from Fig 1 the classification of genes appears to have been defined as follows: the score of gene g≥0.3⇒g is called monoallelic. Otherwise g is called not monoallelic. (Does “not monoallelic” imply biallelicity or do we wish to consider one or more intermediate classes?) However, nothing is explicitly stated in the previous manuscript on the classification of genes independently of individuals. For example (section Results: Landscape of monoallelic expression[…], p6) we read
1b shows the 13 (of 32) genes that did not make it into the top Sg scoring group
but we are not told the criterion based on which the top scoring group was defined.
Other quantities of interest
Although not used for ranking or classification, other fractions were also calculated and presented in Fig 1. First, not only ˆFg(s=0.9) was obtained but also ˆFg(s) for s=0.6,0.7,0.8. Second, the fraction of individuals were calculated that passes the test conceived by Andy: given gene g the fraction of indiv. passed=#{i:sig≤0.6 and UCLig≤0.7}#{i}, where the upper 95 % confidence limit is given by UCLig=sig+z0.975√sig(1−sig)nig, such that zp is the p quantile of the standard normal distribution and nig is the observed total read count.
Results
Genome-wide data import and preparation
## Loading required package: RColorBrewer
Load functions:
source("../../src/import-data.R")
source("../../src/utils.R")
Read count data have been imported (not shown). We see that the minimum number of reads for any gene and individual is 7.
min(unlist(N), na.rm = TRUE)
## [1] 7
The following code
- applies “a filter giving priority to even a single SNP showing biallelic expression” (see Ifat’s ms and her “1_conflict” annotation)
- applies the read count-based filter given a sequence of read count thresholds trc
- applies the individual-based filter given tind
- calculates the fractions of interest, which include the score 1−ˆFg(0.9)
- ranks genes according to their score
min.obs <- 25 # set t_ind
# implementation detail!: filter out genes with fewer observations than 'min.obs'
g.passed <- names(S)[sapply(S, function(y) sum(! is.na(y)) >= min.obs)]
# a sequence of thresholds t_rc for minimum read count to filter individual data points S_{ig} or N_{ig}
min.reads <- c(7, 15, 20)
min.r.names <- paste0("min.reads.", min.reads)
names(min.r.names) <- min.r.names
names(min.reads) <- min.r.names
# filter S_{ig} (then N_{ig}) first based on 'min.reads' and then again on 'min.obs'
Sf <- lapply(min.reads, function(m) filter.min.read(m, X = S[g.passed], N = N[g.passed], min.obs = min.obs))
Nf <- lapply(min.reads, function(m) filter.min.read(m, X = N[g.passed], N = N[g.passed], min.obs = min.obs))
# ECDFs for all filter levels and all genes g; individual ECDF components F_g are named according to gene g
ECDF <- lapply(Sf, sorted.ecdfs)
frac <- lapply(min.r.names,
function(m)
do.fractions(ECDF[[m]], Sf[[m]], Nf[[m]],
frac = 10:6 / 10, ucl.fun = CI.p, max.ucl = 0.7, max.s = 0.6))
Note that the individual-based filter is also applied before the read count-based one. However, this step is an implementation detail and has no conceptual significance.
Result on the individual-based filter
Besides tind=25 the threshold tind=10 was also tested (not shown) but the former resulted in clearly more consistency with Ifat’s ranking (see below).
Gene rankings
The plots show four gene rankings and the corresponding fractions of interest. The first three correspond to the sequence of three trc settings of the read count-based filter so these rankings will be named R7,R15,R20. The fourth plot follows the ranking seen on Ifat’s Fig 1, and this ranking will be referred to as RIfat. Since that figure shows only the 51 genes, the same is done here for also the first three plots. Note that the last 13 genes of the fourth plot are in fact low ranking “known” imprinted genes so they are not in the top 51 according to Rifat. The first three plots do, however, present the 51 top ranking genes for the corresponding ranking.
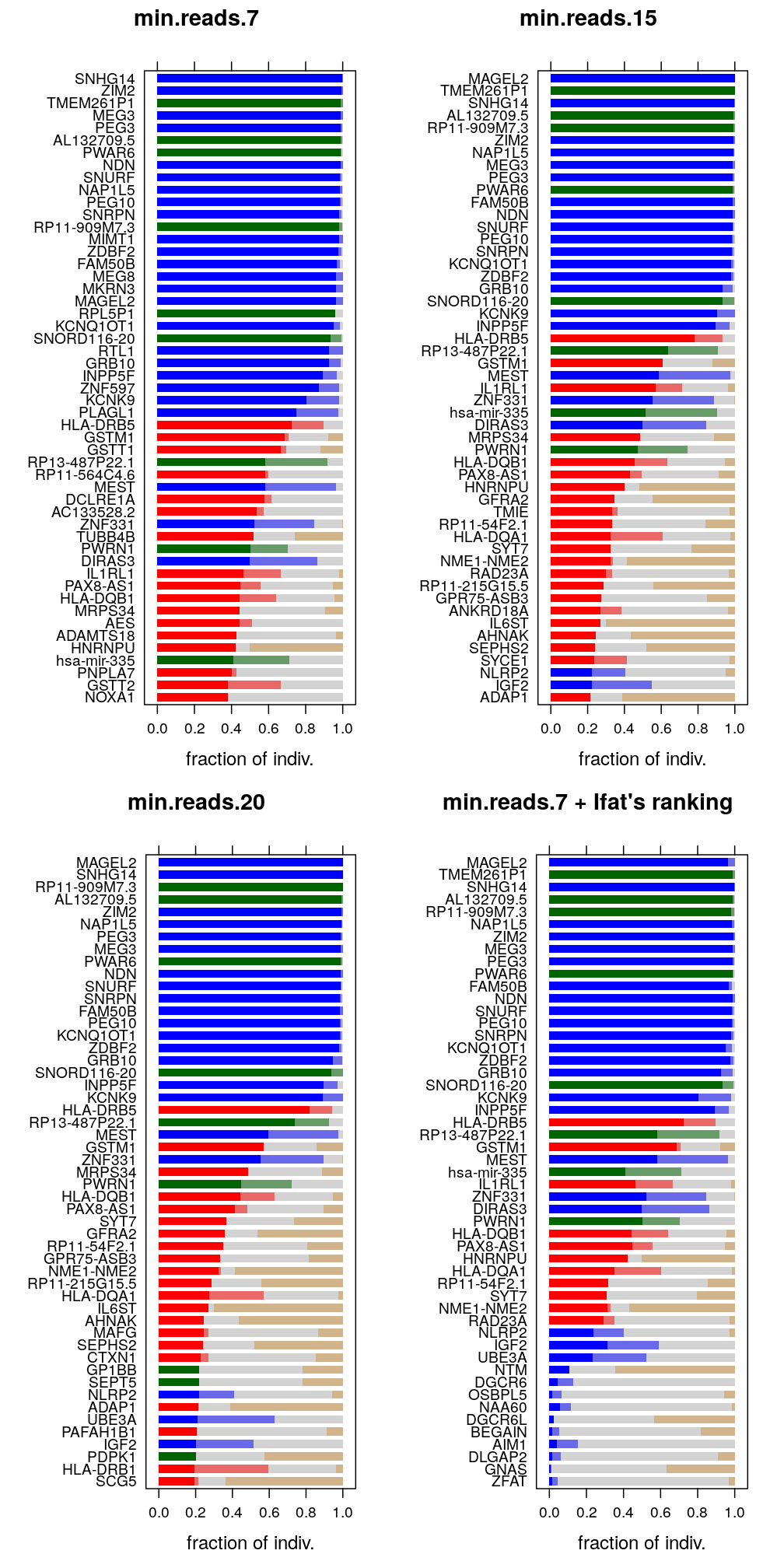
Figure for manuscript
The basis for the figure is the one in the upper-right panel in the previous plot, which is supplemented with the outcome of the “reference/non-reference allele bias” test. The outcome was done via a subjective analysis by Andy and I, in which we sorted each of the top 51 genes into three categories: biased, unbiased, and indeterminate. These are stored in ref-nonref-test.csv and correspond to X, ` ` (whitespace) and 0 in ref.allele.bias, respectively:
ref.allele.bias <- read.csv("../../results/ref-nonref-test.csv")$ref.allele.bias[1:50]
names(ref.allele.bias) <- read.csv("../../results/ref-nonref-test.csv")$gene[1:50]
levels(ref.allele.bias) <- c("X", "0", " ")
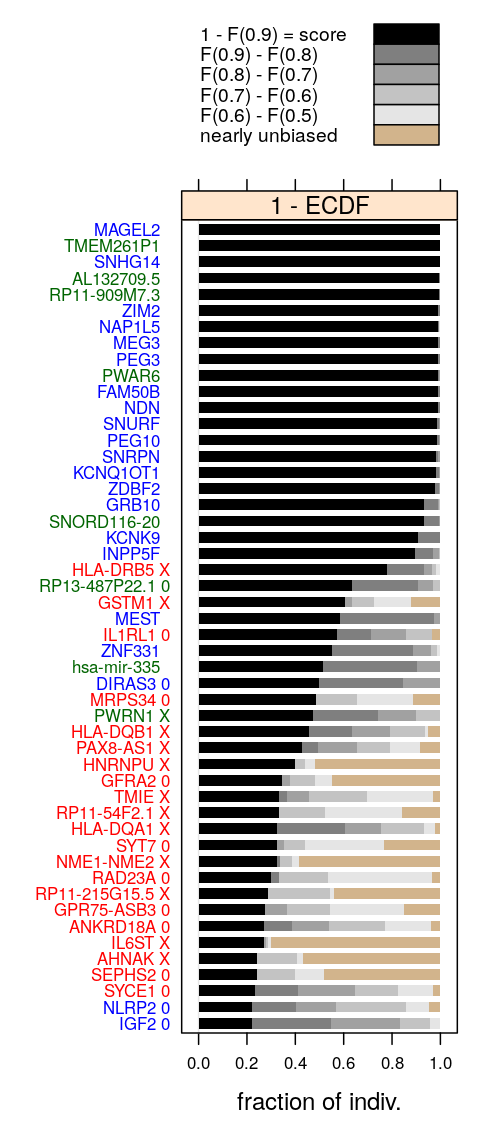
Gene rankings with Ifat’s ranking as reference
The previous figures show that several genes in the top 51 according to R7,R15 or R20 are missing from Ifat’s figure. But clearly, when the read count-based filter is used at trc=15 there is a close match with Ifat’s ranking.
## imprinting.status R.ifat R.7 R.15 R.20
## MAGEL2 known imprinted 1 19 1 1
## TMEM261P1 nearby candidate 2 3 2 NA
## SNHG14 known imprinted 3 1 3 2
## AL132709.5 nearby candidate 4 6 4 4
## RP11-909M7.3 nearby candidate 5 13 5 3
## NAP1L5 known imprinted 6 10 7 6
## ZIM2 known imprinted 7 2 6 5
## MEG3 known imprinted 8 4 8 8
## PEG3 known imprinted 9 5 9 7
## PWAR6 nearby candidate 10 7 10 9
## FAM50B known imprinted 11 16 11 13
## NDN known imprinted 12 8 12 10
## SNURF known imprinted 13 9 13 11
## PEG10 known imprinted 14 11 14 14
## SNRPN known imprinted 15 12 15 12
## KCNQ1OT1 known imprinted 16 21 16 15
## ZDBF2 known imprinted 17 15 17 16
## GRB10 known imprinted 18 24 18 17
## SNORD116-20 nearby candidate 19 22 19 18
## KCNK9 known imprinted 20 27 20 20
## INPP5F known imprinted 21 25 21 19
## HLA-DRB5 distant candidate 22 29 22 21
## RP13-487P22.1 nearby candidate 23 32 23 22
## GSTM1 distant candidate 24 30 24 24
## MEST known imprinted 25 34 25 23
## hsa-mir-335 nearby candidate 26 48 28 NA
## IL1RL1 distant candidate 27 41 26 NA
## ZNF331 known imprinted 28 37 27 25
## DIRAS3 known imprinted 29 40 29 NA
## PWRN1 nearby candidate 30 39 31 27
## HLA-DQB1 distant candidate 31 43 32 28
## PAX8-AS1 distant candidate 32 42 33 29
## HNRNPU distant candidate 33 47 34 NA
## HLA-DQA1 distant candidate 34 57 38 36
## RP11-54F2.1 distant candidate 35 61 37 32
## SYT7 distant candidate 36 65 39 30
## NME1-NME2 distant candidate 37 63 40 34
## RAD23A distant candidate 38 69 41 NA
## NLRP2 known imprinted NA 89 49 44
## IGF2 known imprinted NA 64 50 48
## UBE3A known imprinted NA 92 60 46
## NTM known imprinted NA 335 138 106
## DGCR6 known imprinted NA 1306 646 NA
## OSBPL5 known imprinted NA 3652 987 1009
## NAA60 known imprinted NA 904 1238 1341
## DGCR6L known imprinted NA 2809 1332 1030
## BEGAIN known imprinted NA 4215 1378 1383
## AIM1 known imprinted NA 1534 3600 NA
## DLGAP2 known imprinted NA 3691 3526 3171
## GNAS known imprinted NA 5816 4699 4148
## ZFAT known imprinted NA 3924 4595 4044
In fact, the two rankings agree for the top 24 genes (except that ranks 6 and 7 are swapped):
all.equal(genes.ifat.ranks[ , "R.15"][c(1:5, 7:6, 8:24)], 1:24)
## [1] TRUE
From this result we may conclude that Ifat’s filter settings were trc=15 and tind=25 and the discrepancies we see might partly or entirely be due to rounding errors introduced at the export of S values to csv files and/or different implementation of the calculation of gene scores.
Monoallelically called genes
Given the result that R15 resembles the most to Rifat, we will use R15 to call monoallelic genes. We will compare called gene sets under different threshold for the score 1−ˆFg(0.9) and further compare these to the one presented in the previous manuscript.
Notice that the lowest scoring gene in the “nearby candidate” category is PWRN1 (green on Fig 1). Its score is 0.4714286 and it ranks at 31 according to R15 and at genes.ifat.ranks["PWRN1", "R.ifat"]. We may define the classification threshold such that we only call genes monoallelic if their score ≥0.4:
(called.mono.0.4 <- names(frac$min.reads.15)[unlist(frac$min.reads.15[1, ]) >= 0.4])
## [1] "MAGEL2" "TMEM261P1" "SNHG14" "AL132709.5"
## [5] "RP11-909M7.3" "ZIM2" "NAP1L5" "MEG3"
## [9] "PEG3" "PWAR6" "FAM50B" "NDN"
## [13] "SNURF" "PEG10" "SNRPN" "KCNQ1OT1"
## [17] "ZDBF2" "GRB10" "SNORD116-20" "KCNK9"
## [21] "INPP5F" "HLA-DRB5" "RP13-487P22.1" "GSTM1"
## [25] "MEST" "IL1RL1" "ZNF331" "hsa-mir-335"
## [29] "DIRAS3" "MRPS34" "PWRN1" "HLA-DQB1"
## [33] "PAX8-AS1" "HNRNPU"
If we further lower the classification threshold to 0.3 then according to R15 several more genes must also be called monoallelic.
called.mono.0.3 <- names(frac$min.reads.15)[unlist(frac$min.reads.15[1, ]) >= 0.3]
length(setdiff(called.mono.0.3, called.mono.0.4))
## [1] 7
The upper right panel of the figure above indicates that most—if not all—of the genes between score 0.3 and 0.5 fall in the “distant candidate” category. More on this in the next section.
Implications for regression analysis
My latest regression analysis was carried out on the following genes extending the set of 8 genes initially used in the previous manuscript:
genes.regression.ifat <-
c("PEG3", "INPP5F", "SNRPN", "PWAR6", "ZDBF2", "MEG3", "ZNF331", "GRB10", # 8 genes analyzed by Ifat
"PEG10", "SNHG14", "NAP1L5", "KCNQ1OT1", "MEST", # 5 more genes analyzed by AGK 3/2/16
"IGF2", "NLRP2", "UBE3A", # 3 more genes present in data files
"TMEM261P1", "AL132709.5", "RP11-909M7.3", "SNORD116-20", "RP13-487P22.1", "hsa-mir-335", "PWRN1") # 'green' novel 1 MB imprinted genes; note that PWAR6 is already included above
These were selected based on two rules:
- most monoallelically called genes with imprinting status either “known imprinted” or “nearby candidate”
- exceptions include MAGEL2 and ZIM2—it is not clear why those were not selected initally or later so now they will become selected ones
- the highest scoring but not monoallelically called “known” imprinted genes; these were IGF2, NLRP2 and UBE3A
Given these rules and the called gene sets (at score threshold 0.5 or 0.3) what genes, if any, should I extend my latest (already extended) regression analysis?
I start with the first rule. Excluding “distant candidate”s from the set of monoallelically called genes confirms the earlier suspicion that all genes scoring between 0.3 and 0.4 must be then excluded so for regression analysis it is immaterial which score is used as classfication threshold.
genes.not.cand.4 <- called.mono.0.4[gene.summary[called.mono.0.4, "imprinting.status"] != "distant candidate"]
genes.not.cand.3 <- called.mono.0.3[gene.summary[called.mono.0.3, "imprinting.status"] != "distant candidate"]
all.equal(genes.not.cand.3, genes.not.cand.4)
## [1] TRUE
To see which genes might be selected according to the second rule:
frac$min.reads.15.known <- frac$min.reads.15[gene.summary[names(frac$min.reads.15), "imprinting.status"] == "known imprinted"]
barchart(padded.frac(fr = frac$min.reads.15.known[length(frac$min.reads.15.known):1]),
par.settings = par.set, main = "Known imprinted genes", xlab = xlab)
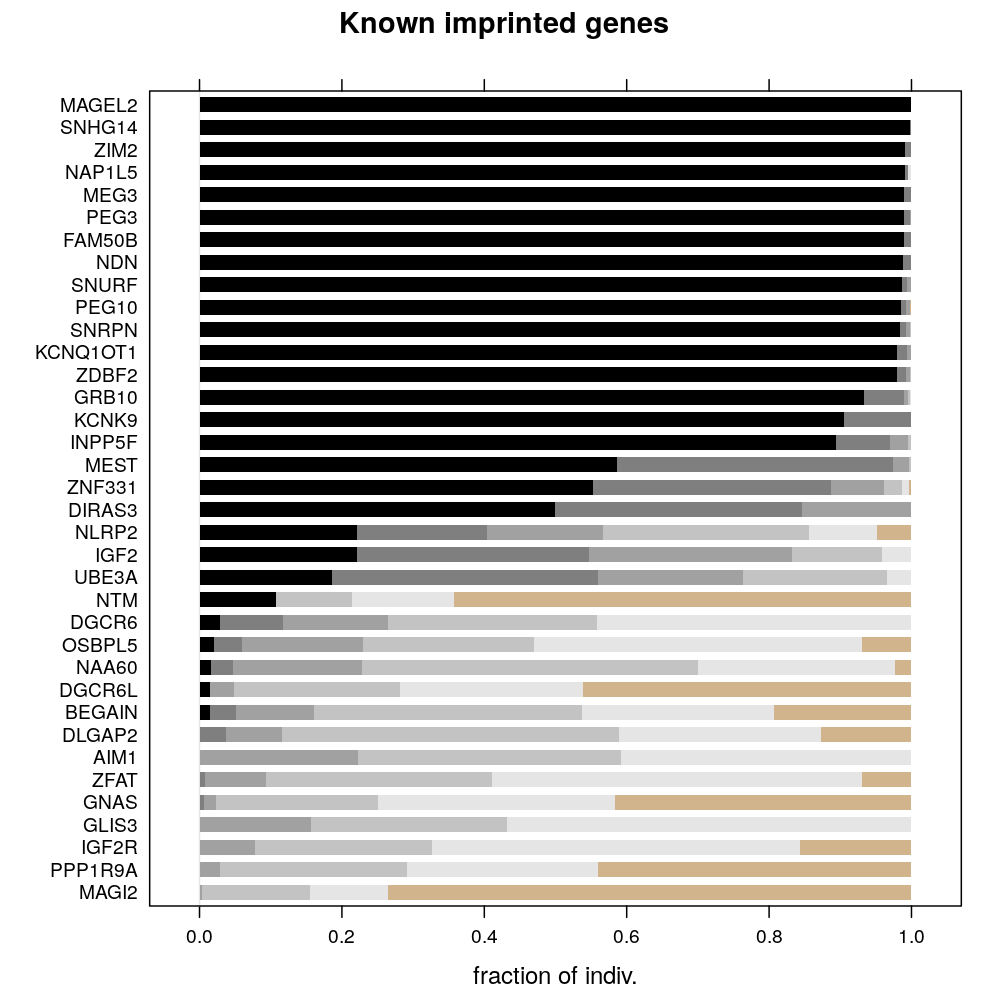
So, it still seems reasonable to select IGF2, NLRP2 and UBE3A based on the second rule.
Then the set of genes to carry out regression analysis:
(genes.regression.new <- c(genes.not.cand.4, c("IGF2", "NLRP2", "UBE3A")))
## [1] "MAGEL2" "TMEM261P1" "SNHG14" "AL132709.5"
## [5] "RP11-909M7.3" "ZIM2" "NAP1L5" "MEG3"
## [9] "PEG3" "PWAR6" "FAM50B" "NDN"
## [13] "SNURF" "PEG10" "SNRPN" "KCNQ1OT1"
## [17] "ZDBF2" "GRB10" "SNORD116-20" "KCNK9"
## [21] "INPP5F" "RP13-487P22.1" "MEST" "ZNF331"
## [25] "hsa-mir-335" "DIRAS3" "PWRN1" "IGF2"
## [29] "NLRP2" "UBE3A"
write.csv(data.frame(genes.regression.new), file = "../../data/genes.regression.new", row.names = FALSE)
No genes need to be removed from the previous set but several new genes need to be added:
setdiff(genes.regression.ifat, genes.regression.new) # what genes to remove?
## character(0)
setdiff(genes.regression.new, genes.regression.ifat) # what genes to add?
## [1] "MAGEL2" "ZIM2" "FAM50B" "NDN" "SNURF" "KCNK9" "DIRAS3"
Cumulative loss of data
The VennDiagram package implements scaled Euler diagrams.
library(VennDiagram)
## Error in library(VennDiagram): there is no package called 'VennDiagram'
The partitions induced by filtering and calling genes monoallelic (imprinted) are illustrated by the following Euler or Venn diagrams. Note that, for an Euler diagram but not for a Venn diagram, the shapes (circles or ellipses) are proportional to the size of the set they represent and that topological relationship among shapes is such that there is no overlap if the intersection of the corresponding sets is the empty set {}.
g.sets <- list(in.dataset = 1:22254, passed.initial.filter = names(S), passed.final.filter = names(Sf$min.reads.15), called.imprinted = genes.regression.new)
sapply(g.sets, length)
## in.dataset passed.initial.filter passed.final.filter
## 22254 15584 5283
## called.imprinted
## 30
l <- c(n.sets <- lapply(g.sets[-4], length), n.sets[-1], rep(n.sets[3], 2))
names(l) <- NULL
l <- c(l, list(category = gsub("\\.", " ", names(g.sets[-4])), cat.cex = 1.2, cat.pos = 0, cat.dist = 1e-2))
grid.draw(do.call(draw.triple.venn, l))
## Error in grid.draw(do.call(draw.triple.venn, l)): could not find function "grid.draw"
## Error in grid.draw(do.call(draw.triple.venn, l)): could not find function "grid.draw"
## Error in grid.draw(do.call(draw.quad.venn, l)): could not find function "grid.draw"
Conclusion
- Ifat’s ranking is the most consistent with the present R15 ranking obtained with filter settings were trc=15 and tind=25
- the consistency is not perfect, which might be due to implementation details
- therefore the two ranking leads to discrepant sets of monoallelically called genes
- the level of discrepancy depends on what rule is chosen for classification
- several considerations play role in what genes are seleceted for regression analysis; these have been discussed above
- the regression analysis needs to be further extended with MAGEL2, ZIM2, FAM50B, NDN, SNURF, KCNK9 and DIRAS3