Imprinted genes form clusters of one or more genes. Previous work by Ifat established the imprinting status of each gene as either known to be imprinted, not known to be imprinted but near an “known” gene, or else neither. I refer to these three categories as “known”, “nearby candidate” and “candidate”, respectively.
The main question queries the mechanism of the age effect on imprinting (loss, gain or lack of effect). In particular: does age regulate genes within some cluster in a concerted or an independent manner?
The current work studies this by performing two main steps
- delineation of clusters
- visualize clusters and age effect in terms of regression coefficients for age
Note that a table comparing imprinted genes identified by Baran et al to those by this study is available at baran-vs-ourwork.csv.
Data import and preparation
## Loading required package: RColorBrewer
Load data importer functions:
source("../../src/import-data.R")
source("../../src/utils.R")
source("~/projects/monoallelic-brain/src/fit-glms.R")
source("2016-08-08-imprinted-gene-clusters.R")
Genome-wide data
gene.summary <-
read.csv("../../data/readcount/summary-all-genes.csv")
rownames(gene.summary) <- gene.summary$Symbol
gene.summary$file.size <-
read.csv("../../data/readcount/fsize-genes.csv", row.names = 2)[rownames(gene.summary), , drop = TRUE]
gene.ids <- with(gene.summary, as.character(Symbol)[ file.size > 0 ])
genes2fit <- unlist(read.csv("../../data/genes.regression.new", as.is = TRUE))
names(genes2fit) <- genes2fit
Y <- get.readcounts(gene.ids, g.subsets = list(), sel.var = c("H", "N"), rm.conflict = TRUE)
S <- data.frame(lapply(gene.ids, function(g) Y[[g]]$H / Y[[g]]$N), check.names = FALSE)
names(S) <- gene.ids
N <- data.frame(lapply(Y, getElement, "N"), check.names = FALSE)
rm(Y)
Filtering
- read count-based filter with threshold trc=15
- individual-based filter with threshold tind=25
The code was copied from an earlier post and is hidden here.
Fitting models to selected genes
Import read count data but do not filter, to be consistent with the most recent regression analysis:
genes2fit <- unlist(read.csv("../../data/genes.regression.new", as.is = TRUE))
names(genes2fit) <- genes2fit
E <- get.predictors() # default arguments
Y <- get.readcounts(gene.ids = genes2fit, count.thrs = 0)
Fitting all models to all retained gene-wise and aggregated read count data sets
# exclude unweighed aggregates UA.8 and UA from fitting
ids2fit <- grep("^UA(.8)?$", names(Y), value = TRUE, invert = TRUE)
M <- do.all.fits(Y[ids2fit], preds = e.vars)
f.ids <- as.data.frame(lapply(M, function(m) ! sapply(m, is.null)))
f.ids["TMEM261P1", c("logi.S", "logi2.S")] <- FALSE
f.ids[c("WA.8", "WA"), ] <- FALSE # gene aggregates are not needed here
Analysis
Delineation of imprinted gene clusters
Let K denote the set of genes g such that g∈K means the gene is either “known” (to be imprinted) or near some “known”, i.e. “nearby candidate”. Let C be the set of “candidate” genes (further away from “known”s). Let gi−1 and gi be neighboring genes on the same chromosome at the i−1-th and i-th site. Delineation of clusters was done using the following simple rule: if gi−1∈C and gi∈K then a cluster starts at the i-th gene. The cluster ends at the first j>i such that gj∈K but gi∈C. The rule is implemented in the make.impr.segs function in 2016-08-08-imprinted-gene-clusters.R.
gene.summary$imprinting.status <- factor(gene.summary$imprinted, ordered = TRUE)
levels(gene.summary$imprinting.status) <- rev(c("known imprinted", "nearby candidate", "distant candidate"))
gene.summary$Symbol <- factor(gene.summary$Symbol, levels = gene.summary$Symbol, ordered = TRUE)
gene.summary$chr <- factor(paste("chr", gene.summary$chr), levels = paste("chr", seq_along(levels(factor(gene.summary$chr)))), ordered = TRUE)
# imprinting segments in component 'seg': clusters and segments inbetween
gs.seg <- make.impr.segs(gene.summary, remove.str = "distant candidate")
gs.seg$cluster <-
factor(x <- with(gs.seg,
ifelse(seg > 0, paste0("clus ", seg, " (", chr, ")"), paste0("inter clus ", abs(seg)))),
levels = unique(x), ordered = TRUE)
rm(x)
# remove filtered genes
gs <- gs.seg[names(frac), ]
gs$score <- unlist(frac["1", ])
Write results to file:
write.csv(gs, file = "../../results/gene-clusters.csv")
Eeach cluster has several genes including the “nearby candidate” category; note the median as well.
cluster.freq <- table(gs.seg$cluster)[seq(2, length(levels(gs.seg$cluster)), by = 2)]
median(cluster.freq)
## [1] 15
barchart(cluster.freq, xlab = "# genes in cluster (including nearby candidates)")
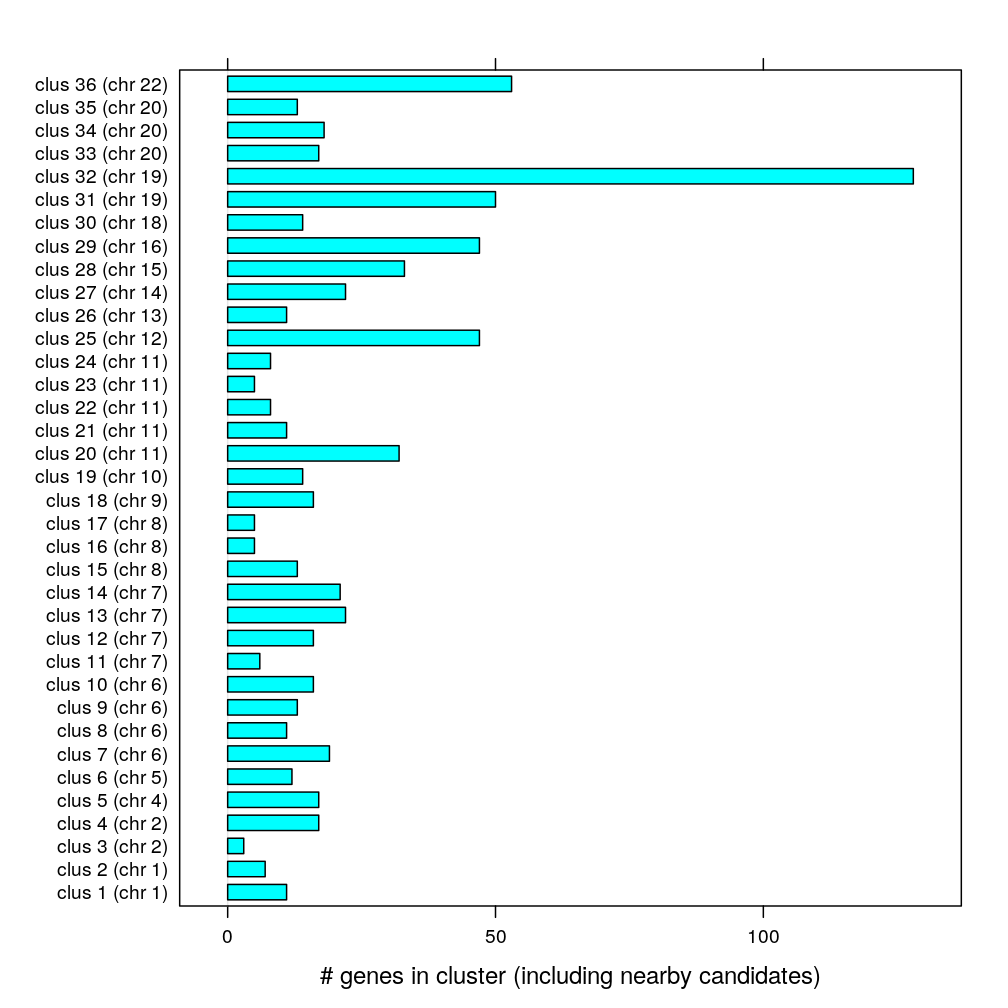
Before filtering these clusters contain the following number of “known” and “nearby candidate” genes:
table(gene.summary$imprinting.status)
##
## distant candidate nearby candidate known imprinted
## 15265 701 60
After filtering:
table(gs$imprinting.status)
##
## distant candidate nearby candidate known imprinted
## 4981 266 36
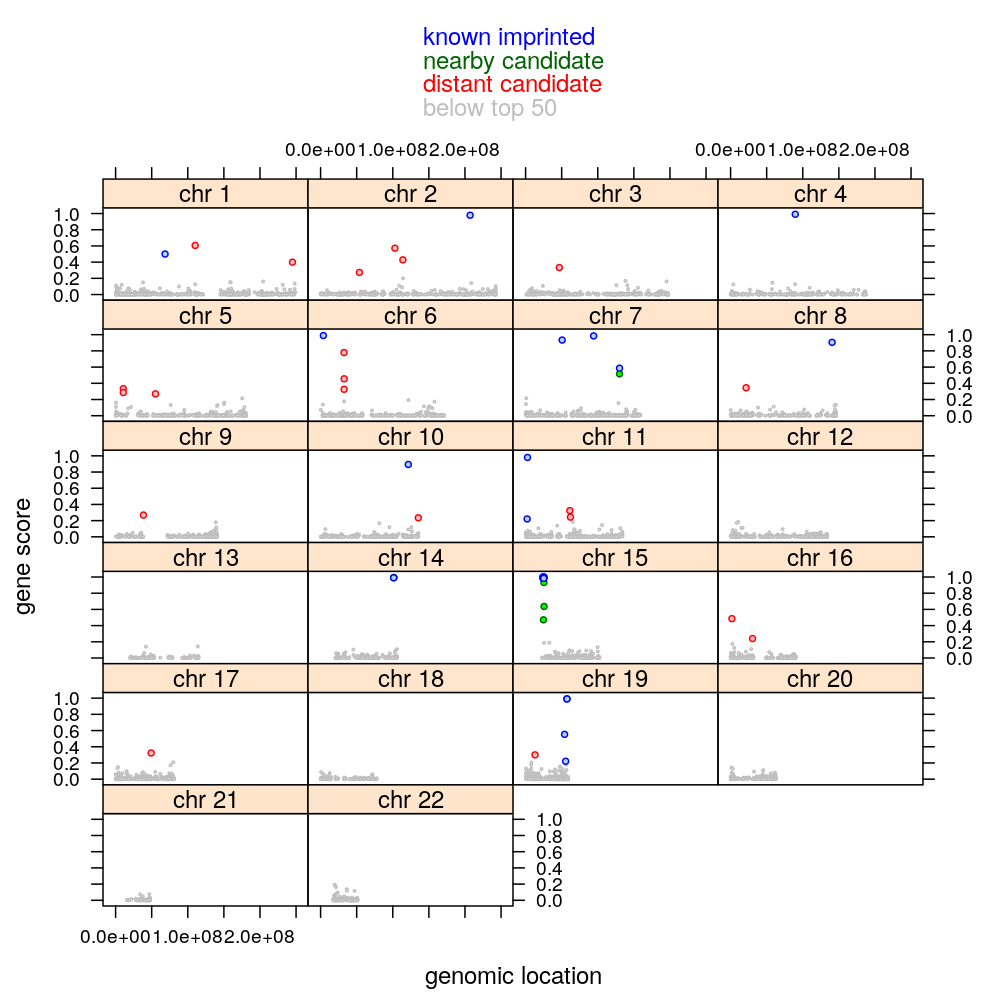
Genomic location
The plot below shows the genomic location of all 5283 genes in the filtered data set and indicates their imprinting status with different colors. Also shown is the gene score according to which genes have been ranked and called as monoallelically expressing or not.
gs$imprinting.status.1 <- as.character(gs$imprinting.status)
gs$imprinting.status.1[51:length(gs$imprinting.status.1)] <- "below top 50"
gs$imprinting.status.1 <- factor(gs$imprinting.status.1, levels = c("below top 50", levels(gs$imprinting.status)))
mycol <- c("gray", "red", "darkgreen", "blue")
xyplot(score ~ start | chr, data = gs, groups = imprinting.status.1,
auto.key = list(text = rev(levels(gs$imprinting.status.1)), columns = 1, col = rev(mycol), points = FALSE),
layout = c(4, 6),
par.settings = list(superpose.symbol =
list(pch = c(21, 21, 21, 21), alpha = c(1, 1, 1, 1),
fill = c("white", "pink", "green", "lightblue"),
cex = c(0.2, 0.5, 0.5, 0.5),
col = mycol)),
xlab = "genomic location", ylab = "gene score")
Extract ˆβage and confidence intervals for βage
sel.genes <- unlist(read.csv("../../data/genes.regression.new", as.is = TRUE))
beta.99 <- lapply(M, function(l.m) do.beta(l.m[sel.genes], conf.lev = 0.99))
beta.95 <- lapply(M, function(l.m) do.beta(l.m[sel.genes], conf.lev = 0.95))
beta.99.long <- do.call(rbind, lapply(names(beta.99), function(x) cbind(beta.99[[x]], data.frame(Model = x))))
#write.csv(beta.99.long, "../../results/beta-99-CI.csv")
beta.95.long <- do.call(rbind, lapply(names(beta.99), function(x) cbind(beta.99[[x]], data.frame(Model = x))))
#write.csv(beta.95.long, "../../results/beta-95-CI.csv")
Filtering for poor fit
Filter based on earlier decisions on the goodness of fit of logi.S, which is stored in results/model-checking.csv.
logi.S.OK <- read.csv("../../results/model-checking.csv", row.names = "gene")["logi.S.fit.OK"]
# set results to NA where logi.S fitted poorly
beta.99$logi.S[beta.99$logi.S$Gene %in% rownames(logi.S.OK)[! logi.S.OK$logi.S.fit.OK],
c("Estimate", "Lower.CL", "Upper.CL")] <- NA
beta.95$logi.S[beta.95$logi.S$Gene %in% rownames(logi.S.OK)[! logi.S.OK$logi.S.fit.OK],
c("Estimate", "Lower.CL", "Upper.CL")] <- NA
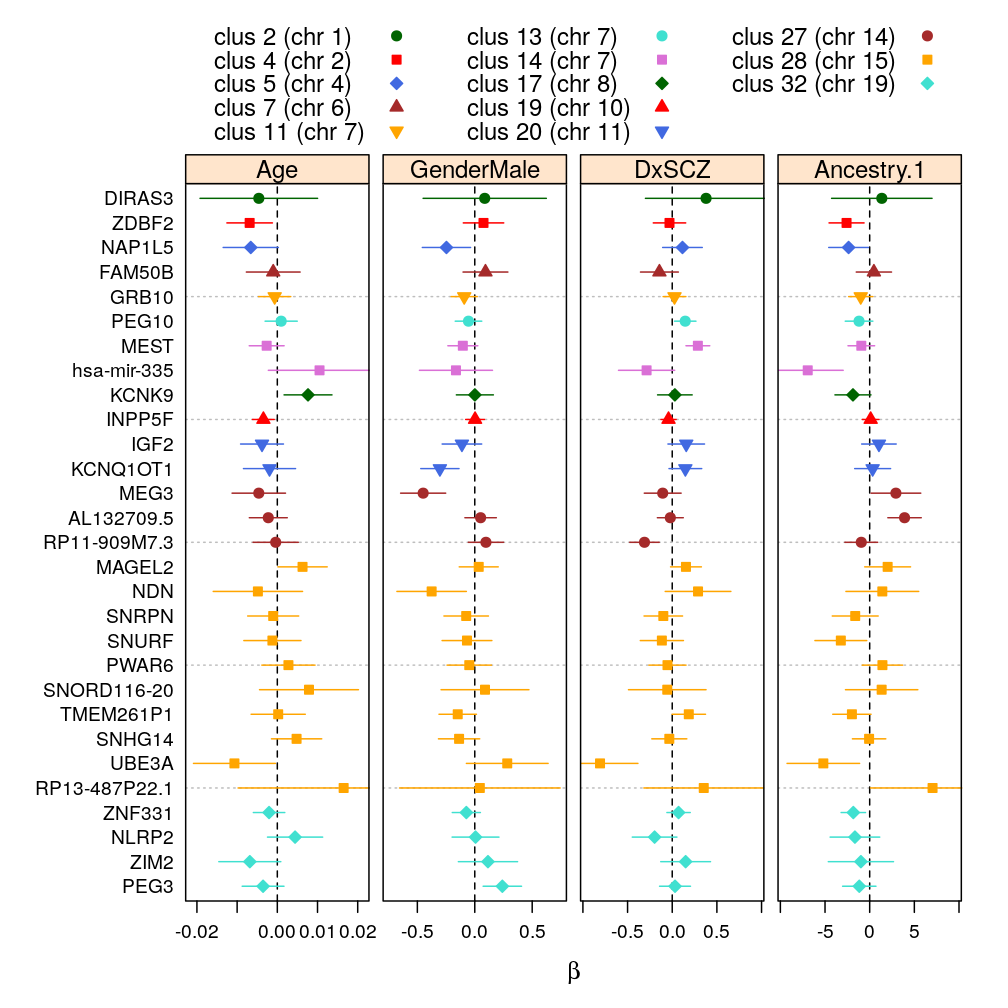
βg arranged by clusters
wnlm.Q, 99 % confidence (for the manuscript)
my.segplot2(beta.99$wnlm.Q, layout = c(4, 1), xlim = list(2e-2*c(-1,1), 0.7*c(-1,1), 0.9*c(-1,1), 9*c(-1,1)))
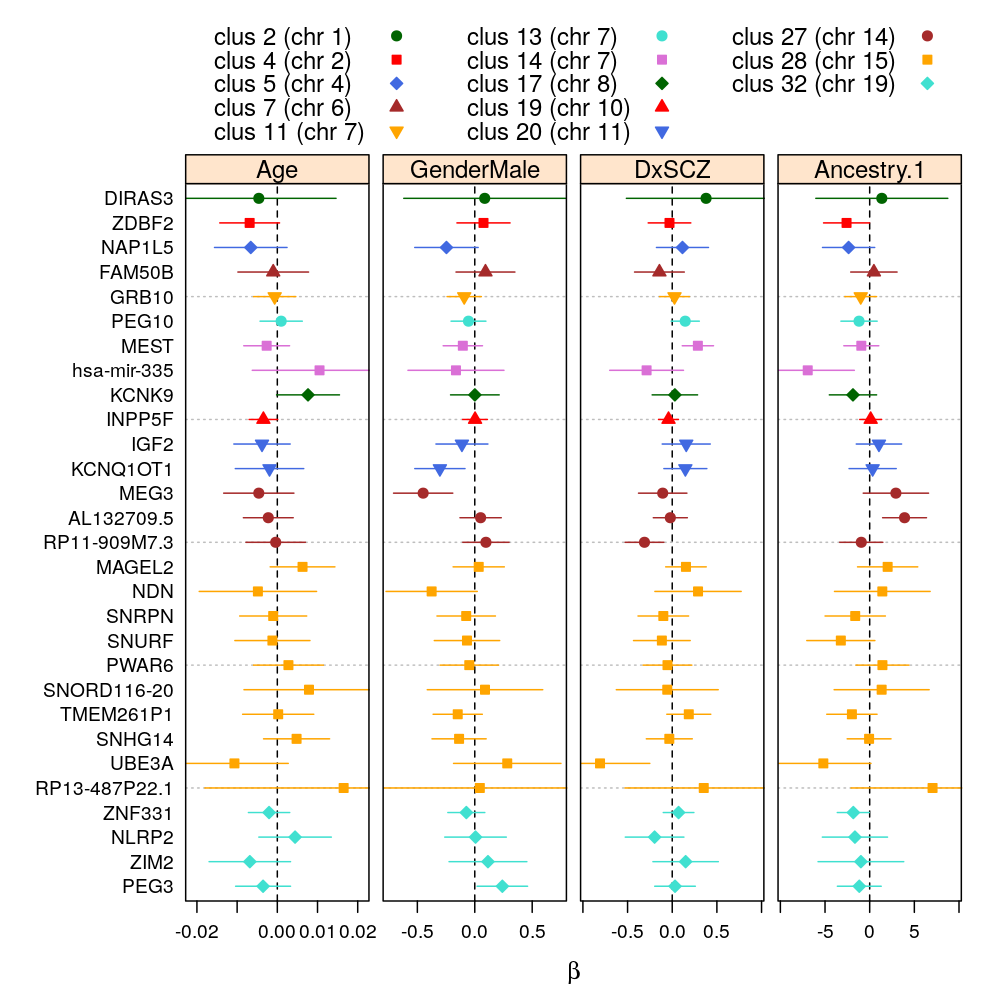
wnlm.Q, 95 % confidence
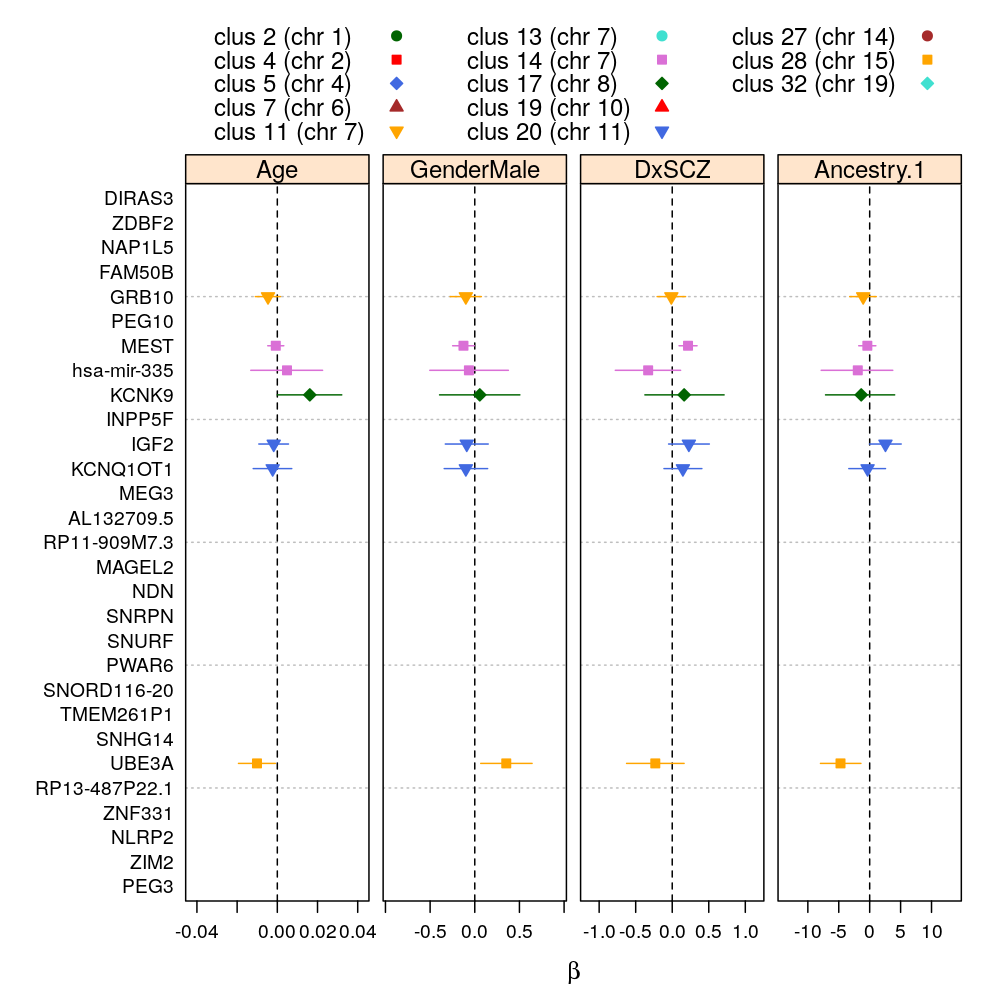
logi.S, 99 % confidence
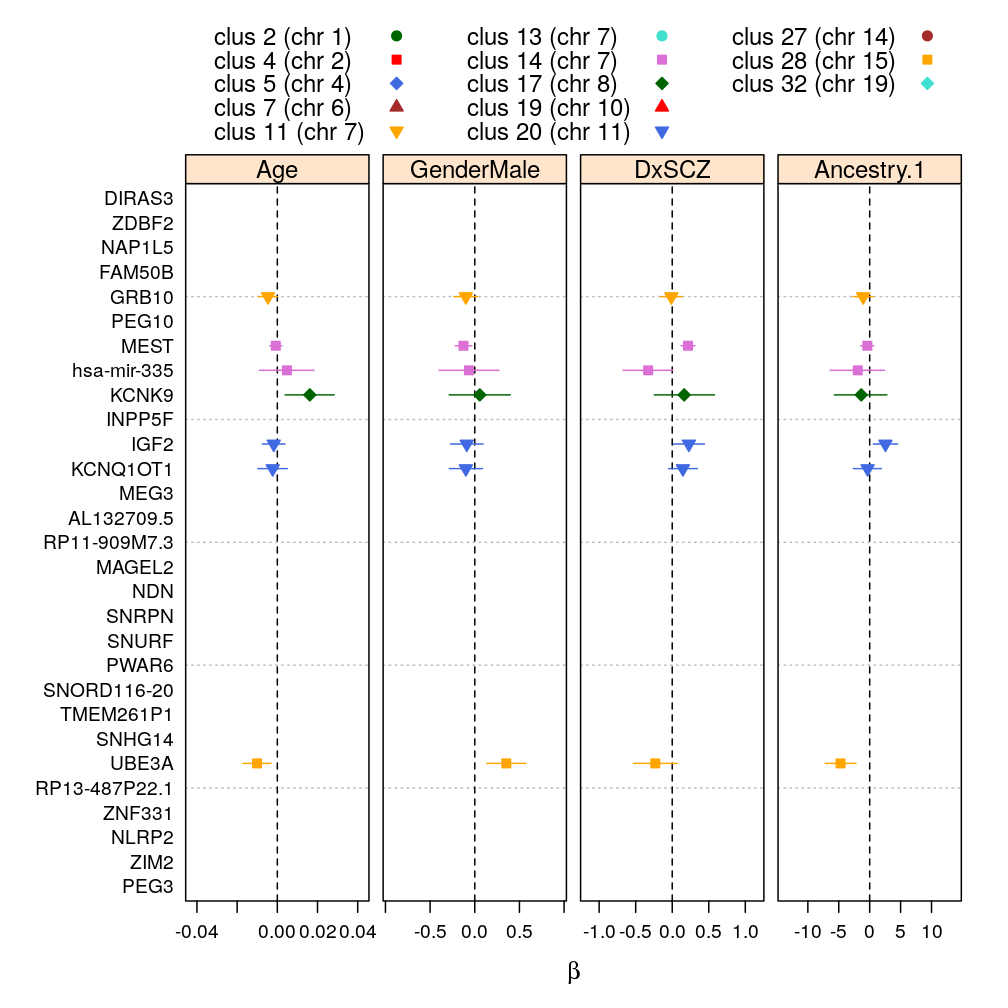
logi.S, 95 % confidence
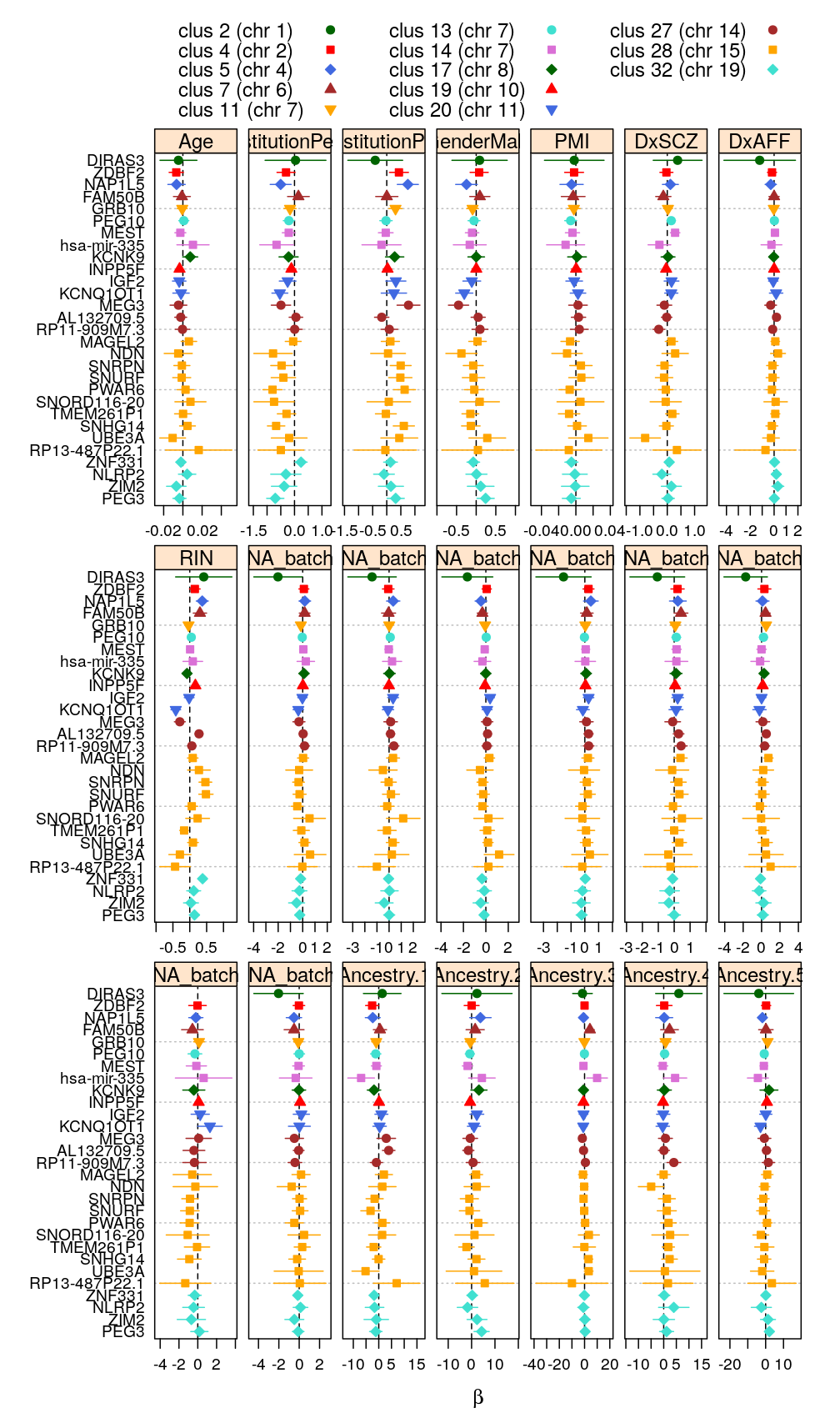
my.segplot2(beta.99$wnlm.Q, layout = c(7, 3), sel.coefs = unlist(lapply(e.vars, function(v) predictor2coefs(M$wnlm.Q[[1]], v))))
Comparison to Baran et al 2015
Baran et al 2015 identified 42 imprinted genes in various human tissues based on GTeX and other data. Their Table S3 lists these genes along with the tissue in which they were found to be imprinted:
baran <- read.csv("../../data/baran-2015-genome-res/table_s3.csv")
levels(baran$name)
## [1] "CPA4" "CST1" "DIRAS3" "DLK1" "FAM50B"
## [6] "GRB10" "H19" "IGF2" "IGF2-AS" "INPP5F_V2"
## [11] "KCNQ1" "KIF25" "L3MBTL1" "LPAR6" "MAGEL2"
## [16] "MAGI2" "MEG3" "MEG8" "MEG9" "MEST"
## [21] "MKRN3" "NAP1L5" "NDN" "NTM" "PEG10"
## [26] "PEG3" "PLAGL1" "PPIEL" "PRSS50" "PWRN1"
## [31] "RP11-7F17.7" "SNHG14" "SNRPN" "SNURF" "SYCE1"
## [36] "THEGL" "UBE3A" "UGT2B4" "UTS2" "ZDBF2"
## [41] "ZNF331" "ZNF597"
Establish consistency of gene names between Baran et al and our study and extract genes Baran et al found to be imprinted in the brain:
levels(baran) <- sub("INPP5F_V2", "INPP5F", levels(baran$name))
levels(baran) <- sub("MEG8", "RP11-909M7.3", levels(baran$name))
baran$name <- sub("INPP5F_V2", "INPP5F", baran$name)
baran$name <- sub("MEG8", "RP11-909M7.3", baran$name)
baran.brain <- baran[ baran$tissue == "BRAIN", "name"]
Summarize genes called imprinted either by Baran et al or this study:
## this.work
## Baran.et.al imprinted not imprinted not assessed
## imprinted 19 12 9
## not imprinted or assessed 10 0 0
Next, list each gene and write result to file. It is noteworthy that the novel imprinted genes suggested by our study are RP11-909M7.3, TMEM261P1, AL132709.5, PWAR6, SNORD116-20, RP13-487P22.1, hsa-mir-335 and among these only RP11-909M7.3 is/are suggested by Baran et al.
baran.vs.ourwork[ , -1]
## Baran.et.al this.work prior.status
## CPA4 imprinted not assessed <NA>
## DIRAS3 imprinted imprinted known_imprinted
## DLK1 imprinted not assessed <NA>
## FAM50B imprinted imprinted known_imprinted
## GRB10 imprinted imprinted known_imprinted
## H19 imprinted not imprinted known_imprinted
## IGF2 imprinted imprinted known_imprinted
## IGF2-AS imprinted not assessed <NA>
## INPP5F imprinted imprinted known_imprinted
## KCNQ1 imprinted not assessed known_imprinted
## KIF25 imprinted not imprinted _
## L3MBTL1 imprinted not imprinted 1 M impr clstr
## LPAR6 imprinted not imprinted 1 M impr clstr
## MAGEL2 imprinted imprinted known_imprinted
## MAGI2 imprinted not imprinted known_imprinted
## MEG3 imprinted imprinted known_imprinted
## RP11-909M7.3 imprinted imprinted 1 M impr clstr
## MEG9 imprinted not imprinted known_imprinted
## MEST imprinted imprinted known_imprinted
## MKRN3 imprinted not imprinted known_imprinted
## NAP1L5 imprinted imprinted known_imprinted
## NDN imprinted imprinted known_imprinted
## NTM imprinted not imprinted known_imprinted
## PEG10 imprinted imprinted known_imprinted
## PEG3 imprinted imprinted known_imprinted
## PLAGL1 imprinted not imprinted known_imprinted
## PPIEL imprinted not assessed <NA>
## PRSS50 imprinted not assessed <NA>
## PWRN1 imprinted not imprinted 1 M impr clstr
## RP11-7F17.7 imprinted not assessed <NA>
## SNHG14 imprinted imprinted known_imprinted
## SNRPN imprinted imprinted known_imprinted
## SNURF imprinted imprinted known_imprinted
## SYCE1 imprinted not imprinted _
## THEGL imprinted not assessed <NA>
## UBE3A imprinted imprinted known_imprinted
## UTS2 imprinted not assessed <NA>
## ZDBF2 imprinted imprinted known_imprinted
## ZNF331 imprinted imprinted known_imprinted
## ZNF597 imprinted not imprinted known_imprinted
## TMEM261P1 not imprinted or assessed imprinted 1 M impr clstr
## AL132709.5 not imprinted or assessed imprinted 1 M impr clstr
## ZIM2 not imprinted or assessed imprinted known_imprinted
## PWAR6 not imprinted or assessed imprinted 1 M impr clstr
## KCNQ1OT1 not imprinted or assessed imprinted known_imprinted
## SNORD116-20 not imprinted or assessed imprinted 1 M impr clstr
## KCNK9 not imprinted or assessed imprinted known_imprinted
## RP13-487P22.1 not imprinted or assessed imprinted 1 M impr clstr
## hsa-mir-335 not imprinted or assessed imprinted 1 M impr clstr
## NLRP2 not imprinted or assessed imprinted known_imprinted
write.csv(baran.vs.ourwork, file = "../../results/baran-vs-ourwork.csv", row.names = FALSE)
Revision for Nature Communications
This part is motivated by the following comments from reviewer #2
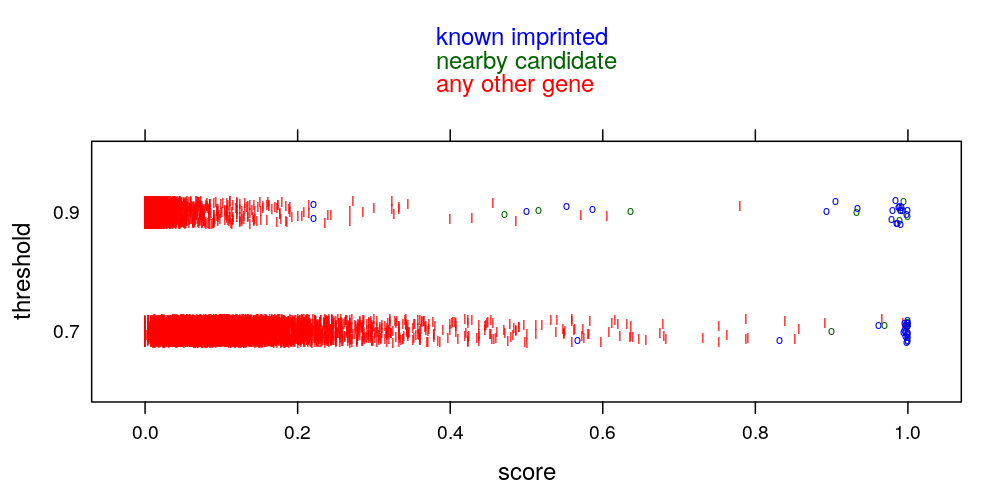
How robust the results are the arbitrarily chosen “S_{ig} > 0.9” threshold? If 0.7 were chosen how would the top 50 genes change? Couldn’t genes simply be ranked by the sum of the S_{ig} scores across individuals? Similar question about the “top 50” genes, why 50, how would the top 20, top 100 behave in terms of imprinting enrichment? Wouldn’t it be simpler to make the point by drawing a ROC curve (truth=known imprinted genes, S_{ig}>.9 fraction as predictor)?
gs.0.9 <- cbind(gs, data.frame(ecdf.thrs = factor("0.9")))
gs.0.7 <- cbind(gs, data.frame(ecdf.thrs = factor("0.7")))
gs$score.0.7 <- gs.0.7$score <- apply(frac[1:3, ], 2, sum)
gs.long <- rbind(gs.0.7, gs.0.9)
rm(gs.0.9, gs.0.7)
mykey <- list(text = c(rev(levels(gs$imprinting.status.1)[-1:-2]), "any other gene"), columns = 1, col = rev(mycol[-1]), points = FALSE, lines = FALSE)
mypar <- list(superpose.line = list(col = c(mycol[-1])), superpose.symbol = list(col = c("red", mycol[-1])))
parallelplot(gs[c("score.0.7", "score")], groups = gs$imprinting.status,
par.settings = mypar, auto.key = mykey, ylab = "threshold", xlab = "score",
scales = list(x = list(at = seq(from = 0, to = 1, by = 0.2), labels = as.character(seq(from = 0, to = 1, by = 0.2))),
y = list(at = 1:2, labels = c("0.7", "0.9"))))
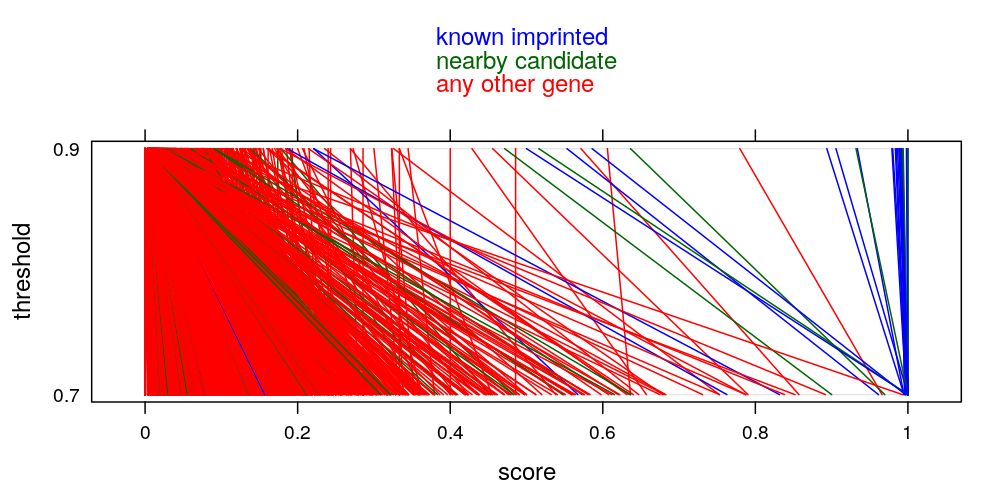
stripplot(ecdf.thrs ~ score, data = gs.long, groups = imprinting.status.1, jitter = TRUE,
par.settings = mypar, ylab = "threshold",
auto.key = mykey, pch = c("I", "I", "o", "o"))